AbstractFull textPDF
Full text
KEY WORDS
Microbiota, hematologic malignancies, antibiotics, immune activation
INTRODUCTION
Overview of the human microbiota: characterization and regulation of microbiota
Human beings are inhabited by innumerable microorganisms that interrelate with the host in a reciprocal way, establishing a combined and efficient ecosystem – the microbiota- that can affect healthiness as well as disease.1,2
The term ‘gut microbiota’ indicates the masses of microorganisms living in the intestine. Most microorganisms inhabit the more distal portions of the intestinal area, where their bio-mass exceeds 1011 cells per gram content.3 Nevertheless, all cavities that connect to the outside and body surfaces are populated by mutable and personalized ecosystems of viruses, fungi, archaea, bacteria and protozoa.
The relationship between the host and the microbiota is symbiotic. The host offers a vital habitat for the microbiome, whereas microorganisms participate in host health via synthesis of essential amino acids, short chain fatty acids (SCFAs) and vitamins.4-6
In a recent study, Sonowal et al. showed that small molecules related to indole and originating from commensal microbiota act in different phyla to augment healthy aging. The action of indoles on health span depends upon the aryl hydrocarbon receptor, a conserved sensor of xenobiotic small molecules. In older animals, indole stimulates genes associated with oogenesis and, accordingly, prolongs reproductive span. These results are improving efforts of developing therapeutics based on microbiota-derived indole to decrease frailty in humans.7
The microbiota is distinguished by its unremitting dynamic renewal of microbiota.8 The different microbiota inhabiting human surfaces are not casually developed. They are the result of several elements, such as age, environmental conditions, lifestyle, smoking habit, antibiotics therapy, genetic factors and contact with pathogenic organisms.9-20 A substantial change in diet modifies microbial configuration within just 24 h of commencement, with a return to baseline two days after diet suspension.21 Moreover, the gut microbiome of animals nourished with a high-sugar or high-fat diet is more disposed to circadian rhythm disruption.22
Numerous diets, including vegan, gluten-free, omnivore, Western and Mediterranean, have been investigated for their ability to regulate the gut microbiota. In numerous studies, a diet high in animal fat and protein, but low in fiber, causes a pronounced reduction in numbers of beneficial Eubacterium species and Bifidobacterium.23-25 Furthermore, host circadian clock and hormonal status alter gut microbial ecology through nourishment and diurnal rhythms; jetlag and long-distance voyage cause the disturbance of this clock, and can therefore alter the gut microbiota.26,27
The initiation of novel tools has strongly impacted the interpretation of the controlling systems by which microorganisms and hosts interrelate to provoke a health or disease condition in the host. Next generation sequencing and methods connected to metabolome analysis, such as mass spectrometry, are critical for evaluating the microbiota structure and investigating the metabolic, functional and genetic action of the microbiota.28-30
Carcinogenesis and microbiota
Oncomicrobes comprise organisms that can directly injure DNA and modify host cellular processes.31-33 Several well-recognized oncomicrobes are viruses, which introduce oncogenes into host genetic material. Interestingly, several bacteria have elaborate competitive tactics which can harm DNA of contending organisms. Unfortunately, these same processes can also modify host DNA, resulting in mutations and perhaps, to carcinogenesis. Bacterial DNA can incorporate into human genomes, principally the mitochondrial genome, through an RNA intermediate, and this occurs more commonly in cancerous rather than healthy tissues.34 DNA mutations may also be caused by toxins generated by bacteria,35,36 and bacterial proteins can initiate signaling actions in host pathways that control cell growth.37,38
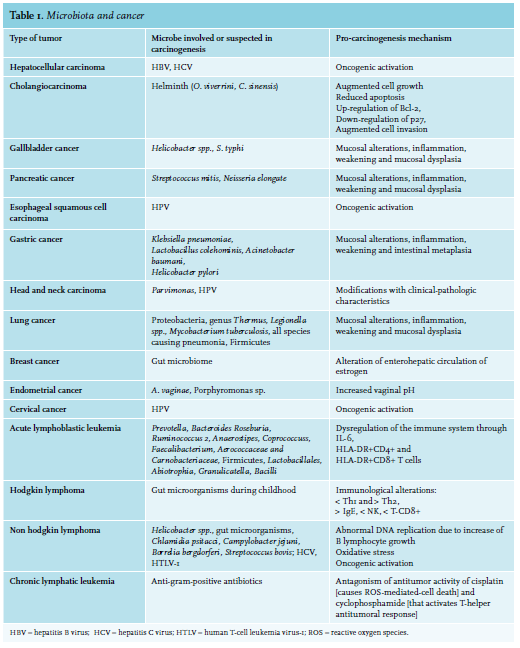 Nevertheless, few microorganisms are identified as oncomicrobes (see table 1). This may partly be due to difficulties in recognizing microorganisms as the causal mediators of carcinogenesis. The causal agent may be absent from the cancer site due to an environmentallydriven population of organisms, or the microbe may have started host cellular injury by a “hit and run” action after only short interaction with the host tissue.
Nevertheless, few microorganisms are identified as oncomicrobes (see table 1). This may partly be due to difficulties in recognizing microorganisms as the causal mediators of carcinogenesis. The causal agent may be absent from the cancer site due to an environmentallydriven population of organisms, or the microbe may have started host cellular injury by a “hit and run” action after only short interaction with the host tissue.
There is increasing evidence that the conformation of the microbiome may influence, and is controlled by, the human immune system.39 An unbalanced microbial structure been connected with reduced immune competence, predisposition to infections and inflammatory diseases.40,41 Experiments performed using germ-free animals propose that microbiota directly stimulate local intestinal immunity through their actions on toll-like receptor (TLR) expression,42 differentiated T cells, antigen presenting cells and lymphoid follicles,43,44 as well as by modifying systemic immunity by augmented systemic antibody production and splenic CD4+ T-cells.45,46
Microorganisms and microbial elements such as lipopolysaccharide (LPS) can up-regulate TLRs, which can provoke an activation of nuclear factor-kB (NF-kB), which is critical for controlling tumor-associated inflammation,47,48 invasion, growth, survival and immunosuppression.49 Bacterial LPS has also been demonstrated to hasten cell proliferation by c-Jun N-terminal Kinase activation.50
Remarkably, T-helper cell 17 (Th17) differentiation from naïve T-cells seems to be dependent on the segmented filamentous bacteria. Experiments have demonstrated that Th17 are lacking in the small-intestinal lamina propria of germ-free mice, which is their primary differentiation location, while modifications in the gut microbiota are closely related to important variations in Th17/Regulatory T-cell (Treg) balance, possibly mediated by epigenetic mechanisms. This is proven by emergent data connecting an unbalanced microbial structure to epigenetic modifications.51-53
Administration with segmented filamentous bacteria provoked an augmentation in production of interferon (IFN) gamma, interleukin-10 (IL-10), and IL-17,54 while administration of Sphingomonas yanoikuyae provoked a modification in immune cells. Bacterioides fragilis induces a Th17 response in animals, which was then demonstrated to be necessary for tumorigenesis, while administration with human commensal bacterium Bifidobacteriom longum, Bacteroides thetaiotaomicron or both caused an increase in the production of the IFN-gamma- and TNF-alpha pathways.55
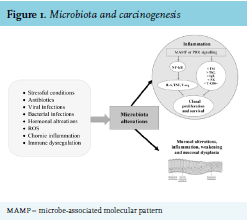 As previously mentioned, gut flora can also control host immunity by epigenetic changes. For example, microbialoriginated butyrate reduces histone deacetylases 6 and 9, which results in higher numbers of Treg cells and enhanced acetylation in the promoter of the FOXP3 gene.56,57 Metabolic relationships and dietary elements can in some way induce tumor expansion. Metabolic end products may include pro-carcinogenic factors that promote cancer growth.58 Contact with these elements, referred to as the “exposome”, can influence oxidative stress and DNA stability in a host, increasing the danger of developing tumor.59,60 (see figure 1). Several carcinogenic factors can be produced by the cometabolism of xenobiotics by bacterial β-glucuronidases.61 Significant examples comprise the metabolism of azoxymethane62 and irinotecan.63 Similarly, production of harmful metabolites is connected with microbial catabolism of dietary proteins. These putrefactive activities in the bowel provoke the production of N-nitroso elements that cause DNA damage via alkylation.64,65 The metabolism of aromatic amino acids also causes the production of phenols, indoles, p-cresol and phenylacetic acid.66 Polyamines are a diverse group of toxic elements and catabolism of the main polyamines is connected with oxidative stress and tumors67 (see table 2).
As previously mentioned, gut flora can also control host immunity by epigenetic changes. For example, microbialoriginated butyrate reduces histone deacetylases 6 and 9, which results in higher numbers of Treg cells and enhanced acetylation in the promoter of the FOXP3 gene.56,57 Metabolic relationships and dietary elements can in some way induce tumor expansion. Metabolic end products may include pro-carcinogenic factors that promote cancer growth.58 Contact with these elements, referred to as the “exposome”, can influence oxidative stress and DNA stability in a host, increasing the danger of developing tumor.59,60 (see figure 1). Several carcinogenic factors can be produced by the cometabolism of xenobiotics by bacterial β-glucuronidases.61 Significant examples comprise the metabolism of azoxymethane62 and irinotecan.63 Similarly, production of harmful metabolites is connected with microbial catabolism of dietary proteins. These putrefactive activities in the bowel provoke the production of N-nitroso elements that cause DNA damage via alkylation.64,65 The metabolism of aromatic amino acids also causes the production of phenols, indoles, p-cresol and phenylacetic acid.66 Polyamines are a diverse group of toxic elements and catabolism of the main polyamines is connected with oxidative stress and tumors67 (see table 2). 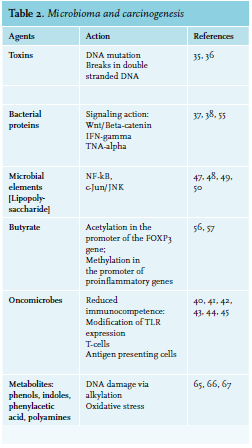
Chemotherapy and microbiome
Chemotherapy can harm normal cells of the intestinal system and possibly cause gastro-Intestinal (GI) problems.68 The cytotoxic actions of these therapies provoke a supplementary immunosuppression, which causes febrile neutropenia and bloodstream infections. Furthermore, the usage of prophylactic and therapeutic antibiotics alters the GI microbiome.69
Galloway-Peña et al. studied intra-patient temporal microbiota changeability and its clinical impact on tumor subjects during chemotherapy.70 These patients presented an elevated level of intra-subject temporal instability of oral and fecal bacterial multiplicity. Days on antibiotics were substantially connected with extended temporal variability of oral bacterial multiplicity and microbiome structure. These results indicate that an increased variability was connected with adverse clinical prognosis. Moreover, the data show the relevance of longitudinal microbiome analyses.
Unfortunately, direct interaction with bacteria can influence the effectiveness of chemotherapeutic agents.71 Mass spectrometry and high-performance liquid chromatography analysis demonstrated that contact with the bacteria determined a biotransformation of certain chemotherapy drugs. The ability of bacteria to reduce the antitumor effect of gemcitabine and to increase that of the prodrug CB1954 was demonstrated in vivo.72 Hence, data in experimental models propose a complicated interaction between numerous chemotherapeutic agents and microbiota.
Exhaustive mechanistic studies in vivo have been reported for platinum compounds and cyclophosphamide (CTX).73,74 CTX, prescribed for therapy of hematologic diseases and solid cancers, harms the small intestine epithelium causing migration of several microorganisms into lymph organs. This barrier breech provokes a T-helper cell-mediated anticancer response and augments drug effectiveness.73 The antitumor action of platin treatment is radically reduced in germ-free mice or in mice in which gut bacteria have been reduced by broad-spectrum antibiotics.74 Combined with experimental results that Lactobacillus acidophilus supports the anticancer action of cisplatin, clinical data propose the possibility that probiotic bacterial species may promote antitumor action while inhibiting some of the toxic side effects of particular drugs75-77 (see table 3). 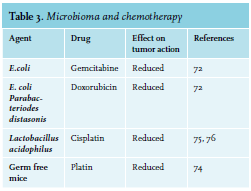 The intestinal chemotoxicity of methotrexate is due in part, by activation of TLR4 by microbial products such as Cif, as secreted toxin from Pseudomonas aeruginosa. Activation of TLR2 protects the mucosa against methotrexate-induced damage by augmenting the expression of the ABC transporter multidrug resistance protein 1 (MDR1; also known as P -glycoprotein and ABCB1), which controls the efflux of xenobiotics from intestinal epithelial cells.78
The intestinal chemotoxicity of methotrexate is due in part, by activation of TLR4 by microbial products such as Cif, as secreted toxin from Pseudomonas aeruginosa. Activation of TLR2 protects the mucosa against methotrexate-induced damage by augmenting the expression of the ABC transporter multidrug resistance protein 1 (MDR1; also known as P -glycoprotein and ABCB1), which controls the efflux of xenobiotics from intestinal epithelial cells.78
Microbiota and immunotherapy
Improvements in treatments and immunotherapies have generally discounted microorganisms as a part of the tumor therapy, until recently. The intestinal microbiome has been discovered to modify responses to tumor immunotherapy.79
Vetizou et al.80 and Sivan et al.81 demonstrated that the efficiency of immune checkpoint inhibitors (ICIs) in therapy is reliant on the host microbiota and that the ICIs responded poorly in animals raised under germ-free situations. The authors discovered that, in the presence of microorganisms, host antigen-presenting cells stimulate IFN γ-generating T-cells. Vetizou et al. studied how the curative efficiency of cytotoxic T-lymphocyte-associated protein 4 blockade, as mediated by ipilimumab, might also be caused by elements of the gut microbiota.82
It is now clear that primary resistance to immune checkpoints can be attributed to an abnormal gut microbiome composition. Fecal microbiota transplantation (FMT) from cancer patients who responded to immune checkpoints into germ-free or antibiotic-treated mice ameliorated the antitumor effects of programmed cell death 1 protein (PD-1) blockade, whereas FMT from non-responding patients failed to do so. Oral supplementation with Akkermansia. muciniphila after FMT with non-responder feces restored the efficacy of PD-1 blockade in an IL-12-dependent manner by increasing the recruitment of CCR9+CXCR3+CD4+ T lymphocytes into mouse tumor beds.83 Moreover, in melanoma patients undergoing anti PD-1 immunotherapy, significant differences were observed in the diversity and composition of the patient gut microbiome of responders versus non-responders. Analysis of patient fecal microbiome samples showed significantly higher alpha diversity and relative abundance of bacteria in the Ruminococcaceae family in responding patients.84
Microbiota and bone marrow transplantation
Hematopoietic stem cell transplantation (HSCT) is frequently used as curative therapy in patients with hematological diseases. The microbiome could be implicated in local and systemic complications after HSCT. Antibiotic-caused loss of lower gut diversity has been suggested as an independent predictor of unfavorable outcome, while progress of graft-versus-host disease (GVHD) appears to be a main contributor to mortality.85,86 Lately, microorganisms of the genus Blautia were identified as the connection between modifications in microbiota structure, onset of GVHD and a favorable outcome.87,88 In fact, increased amounts of bacteria belonging to the genus Blautia were associated with reduced GVHD lethality. Blautia abundance was also associated with improved overall survival.87 The role of the intestinal microbiota and its potential influence on clinical outcomes for patients undergoing allogeneic HCT (allo-HCT) has been investigated in recent years.89-91
Acute GVHD (aGVHD) might reduce the chances of successful allogeneic bone marrow transplantation. Present pathophysiologic hypotheses of aGVHD include proinflammatory cytokines and bacterial LPS as main triggers for aGVHD. LPS originates principally from Gram-negative bacteria and can pass in circulation via the damaged mucosal barrier after the conditioning regimen. Probiotic microorganisms have been proven to modify the components of the intestinal microflora and thus mediate anti-inflammatory actions. It has been suggested that changing the enteric flora using the probiotic microorganism Lactobacillus rhamnosus GG would improve aGVHD.92,93 Analyses of fecal specimens taken from recipients of allo-HCT around the time of engraftment have shown that a reduced intestinal microbiome diversity is associated with significantly worse survival outcomes, and that the alterations in the microbiota composition may influence important clinical outcomes such as aGVHD and disease relapse.94-96
It has been hypothesized that approaches to restore a patient’s microbiome diversity after HCT may improve outcomes after HCT. Although approaches in restoring microbiome diversity are still investigational, FMT from a healthy individual promise to be a remarkably effective therapy.97 FMT refers to the infusion of fecal suspension from a healthy donor into the gastrointestinal tract of a patient in order to restore the microbiota and cure the disease. Manipulation of the intestinal microbiota by FMT may influence the immune system and improve immune-mediated enteritis such as gut aGVHD.98
Moreover, bloodstream infection (BSI) is one of the biggest causes of death among cancer patients. A study stated that gut domination, described as the occupation of at least 30% by a specific bacterial taxon, is connected with BSI in subjects undergoing allo-HSCT. These data suggest that the intestinal microbiota can recognize high-risk subjects before HSCT and that management of the intestinal microbiota to prevent BSI in high-risk subjects could be a useful treatment.99
Finally, a common side effect of myeloablative therapy used during HSCT treatment is GI mucositis.100 A model, created by Sonis, illustrated a process for bacterial infection due to GI mucositis after HSCT.101 It comprises an ulcerative stage with increased permeability and harm to the gut mucosal barrier. This stimulates microbial translocation, described as the migration ofmicroorganisms from the GI tract to extra-intestinal places, such as the blood.102
Antibiotics, microbiota and cancer
The discovery that a persistent usage of antibiotics can stimulate some tumors in cancers such as esophageal, gastric and pancreatic cancers,103 suggests an association between antibiotics, microbiota and cancer.
In fact, there is an indirect indication that an unbalanced microbial structure provoked by antibiotic treatment can augment the incidence of some tumors. An epidemiological study (125,441 cases and 490,510 matched controls) suggests that the use of antibiotics may also influence the occurrence of breast cancer (macrolides, penicillin, cephalosporins, sulphonamides and tetracyclines).103,104 These data propose that some antibiotics are carcinogenic (tetracyclines inhibit replication of mitochondrial DNA) or that they cause changes in the structure of the microbiota that promote the growth of cancers. However, these results have to be cautiously interpreted because they referred to solid tumors, and the iterative use of antibiotics may denote the existence of immune alterations that could be the main cause of microorganism infections and increased tumor frequency.
Nevertheless, antibiotics can enhance the growth of tumors in animals. This has been demonstrated for the therapy of proto-neu transgenic animals in which tumors appear with ciprofloxacin plus metronidazole.105,106
Diet, probiotics and microbiota
There is outstanding interest in the manipulation of the microbiome as a possibility to reduce the incidence of cancer. The intrinsic relationship between microbiome, diet and health demonstrates a possible improvement of our health if we modify our diet.107,108 Indeed, the American Cancer Society has indicated that diets might be responsible for 30% of tumor cases in developed states and 20% in developing nations.109
Reports indicate that diet modifies the structure of the gut flora, and this can impact on the immune response.110,111 On the other hand, eating particular types of foods (fish, fruits, poultry and vegetables) may prevent several tumors.112-115
Metabolism of food can also cause the production of bioactive molecules with chemo-protective activities. Carbohydrate fermentation provokes the production of the short chain fatty acids acetate, butyrate and propionate. These substances can act with free fatty acid receptors in the gut epithelia to influence immune processes, such as the production of cytokines.116 Other researchers have demonstrated that butyrate can increase the number of formations of tight junction proteins through the modulation of AMP-activated protein kinase.117 Butyrate also has anti-carcinogenesis actions, principally by two means: the stimulation of G-protein-coupled receptors 41 and 43 and the reduction of histone deacetylase. Some of the described actions of butyrate are improvements of specific pro-apoptotic gene expression in tumor cells and the reduction of the pro-inflammatory pathway of NF-kB.118,119
Moreover, there is epidemiological evidence supporting chemo-protection derived from the intake of vegetables and fruits, which have been ascribed to secondary metabolites, such as polyphenols and glucosinolates.120 Polyphenols are powerful anti-oxidants, but experiments have demonstrated that they are not able to achieve bioactive levels in the blood.121,122 These elements can also modulate immune responses by modification of the intestinal microbial structure.123,124
Probiotics can live in the gut and stimulate the recuperation of regular intestinal microbiota; the most well-known probiotics are Bifidobacteria and Lactobacillus. 125
Probiotics increase the immune response through augment of natural killer cell (NK) cytotoxicity and the stimulation of phagocytes.126 The effectiveness of NK cells can also be augmented when a mixture of probiotics and dextran is utilized.127 Furthermore, the increase of probiotics seems to increase the production of immunoglobulins IgM and IgA, thus fortifying the adaptive immune response.128-132
Bifidobacteria and Lactic acid bacteria are able to induce the secretion of elements that reduce inflammation by downregulating IL-8 production, NF-kB-dependent gene expression and concentrations of macrophage-attracting cytokines.133-136 Morita et al. also demonstrated a significant augmentation of the expression of IL-6, IL-10, and IL-12 after the stimulation of macrophages with Lactobacillus acidophilus.137 Nutritional supplements with probiotics have been used to increase immune system activity in elderly people.138,139 Li et al. showed that Prohep - a new probiotic mix - reduces cancer development in an animal model.140 Probiotics change the intestinal bacterial structure so that it comprises specific advantageous microbes, such that Oscillibacter and Prevotella, known fabricators of anti-inflammatory substances, which are able to decrease Th17 polarization and stimulate the differentiation of Treg/ Tr1 cells in the intestine.
Microbiota and hematologic malignancies
Acute Lymphoblastic Leukemia (ALL)
Rajagopala et al. compared the GI microbiota constitution of adolescent and pediatric leukemia subjects with their healthy relatives.141 They identified modifications in the microbiota composition of leukemia subjects during chemotherapy by evaluating samples taken before and after treatment at variable time points throughout the treatment. Their results supply relevant data on GI microbiota structure in immunocompromised patients and suggest that the baseline microbiota of these patients is significantly different from their healthy siblings.
The microbiota structure of patients and siblings are dominated by components of Prevotella, Bacteroides and Faecalibacterium. The microbiota diversity of the patient groups was substantially lower than that of the controls. It was possible to differentiate between the leukemia subjects and the controls based on their microbiota composition. The principal taxa comprise Roseburia, Ruminococcus2, Anaerostipes and Coprococcus with moderately higher abundance in the controls.141
Information regarding the oral microbiota in leukemia subjects is lacking and mostly inadequate. Among some patients, leukemia first presents in the oral cavity.142,143 Oral symptoms that commonly arise in leukemia subjects include gingival enlargement and bleeding, candidiasis, oral ulceration and periodontitis144-146 and oral microorganisms are implicated in the onset of such problems.147 Specific oral microorganisms have been shown to contribute to septicemia, which might delay antineoplastic therapy or even put the subjects’ life at risk.148-153 Consequently, a suitable therapy for oral lesions could result in a more satisfactory outcome of both oral and systemic complications.
However, Wang et al. studied the structure of the supragingival plaque microbiota of ALL pediatric subjects and of the healthy controls.154 The oral microbiota of leukemia subjects had less diversity related to controls. Microorganisms grouped into two main clusters, patients and controls, with diverse composition. Variations of specific taxa comprising the families Aerococcaceae and Carnobacteriaceae, Phylum Firmicutes, the order Lactobacillales, the genera Abiotrophia and Granulicatella and the class Bacilli were correlated with leukemia status. Nevertheless, it was demonstrated that the complexity of oral microbiota was not significantly diverse between leukemia subjects and controls until beginning of antineoplastic therapy. This contradiction might be ascribed to the methods employed to study oral microbiota. At the time of the study, oral microorganisms might have already undergone selection after ALL was detected, with some microorganisms inhibited and others prospering.155
However, the studies of oral microorganisms in ALL subjects, however, provided the possibility of recognizing potential microorganisms correlated with systemic infections in leukemia subjects. Results propose two taxonomical lineages (Firmicutes/Bacilli/Lactobacillales/Aerococcaceae/Abiotrophia, and Firmicutes/ Bacilli/Lactobacillales/Carnobacteriaceae/Granullicatella) that are much more copious in the supragingival plaque of ALL subjects than controls. This suggests that advantageous situations existed for their growth in the oral space of ALL subjects and could be responsible for an increased risk of bacteremia in leukemia subjects.
Abiotrophia and Granulicatella have been involved in endocarditis, otitis media, central nervous system infections, cholangitis and arthritis.156-159 Previous researchers have found that oral microorganisms are responsible for local infections and for 25-50% of systemic infections.160
Finally, adult survivors of ALL have health problems that arise years after termination of treatment. Chua et al. evaluated the anal microbiota structure of adult survivors of childhood ALL and controls. They recognized a modified population with decreased microbial diversity in tumor survivors, who also display signals of immune alteration comprising enhanced T-cell activation. The microorganism population among ALL survivors was enriched for Actinobacteria and depleted of Faecalibacterium, consistent with corresponding plasma levels of C-reactive protein and IL-6 and HLA-DR+CD4+ and HLA-DR+CD8+ T cells. They established a relationship between dysbiosis and immune alteration in adult ALL survivors. Actions that could reestablish microbial diversity may improve development of late effects of childhood ALL survivors, particularly chronic inflammation-related comorbidities.161
Acute myeloid leukemia (AML)
Bacterial infections and their complications are one of the most frequent and critical treatment-related toxicities in subjects with AML. Gram-positive cocci, principally the heterogeneous Viridans streptococci, are the most usually isolated microorganisms in patients with AML.162 However, many elements of the microbiota could play a positive role towards leukemic disease. The oral bacterium, Aggregatibacter actinomycetem comitans, generates a leukotoxin (LtxA) that is specific for white blood cells by interacting with lymphocyte function antigen-1 (LFA-1) on susceptible cells. Kachlany et al. valuated the in vitro and in vivo anti-leukemia action of the toxin. LtxA destroys malignant cell lines and primary leukemia cells from AML subjects, while healthy cells are moderately resistant to LtxA-mediated cytotoxicity. Levels of LFA-1 in Jurkat cell lines correlated with killing by LtxA and the toxin especially destroyed cells presenting the activated form of LFA-1. In a severe combined immune deficiency mouse model for human leukemia, LtxA had powerful therapeutic action resulting in long-term survival of the LtxA-treated mice.163
Interestingly for patients with erythroleukemia, a rare form of AML, kefir, a beverage obtained by the incubation of kefir grains with raw milk may be an effective therapy. Kefir grains are a symbiotic complex of diverse kinds of yeasts and bacteria, especially lactic acid bacteria, which congregate in a mostly carbohydrate matrix, called kefiran. In recent years, the action of kefir on some cancers has been investigated. Jalali et al. demonstrated that kefir caused apoptosis and necrosis in an acute erythroleukemia cell line (KG-1), by reducing growth. The study suggested that kefir may have the potential to be an effective therapy for erythroleukemia.164
Furthermore, numerous bacterial toxins are being investigated as potential anti-leukemia agents, either for their direct effects or to release therapeutic proteins against leukemia. LukS-PV, an element of Panton-Valentine leukocidin (PVL) produced by S. aureus, has certain anti-leukemia actions such as inducing leukemia cell differentiation and apoptosis, thus making LukS-PV an encouraging novel treatment strategy for leukemia.165 PVL is a staphylococcal synergohymenotropic exotoxin belonging to the pore-forming toxin family. PVL causes lysis of human polymorphonuclear neutrophils, monocytes and macrophages. Several works have proven that LukS-PV is able to cause leukemia cell differentiation.166 LukS-PV provoked differentiation by stimulating the extracellular-signal-reduced kinase (ERK) signaling pathway and c-JUN/c-FOS in human acute myeloid leukemia cells.167,168
B and T lymphomas
Adolescent/young adult Hodgkin lymphoma (AYAHL) is associated with reduced exposures to infections. Similarly, a study of AYAHL survivors suggested fewer early childhood fecal-oral exposures compared with health controls, and patients have immunological alterations. AYAHL is related to suppressed Th1 activity and an increase of Th2 response.169
Extension of gut microorganisms during childhood170,171 correspond with a change from a Th2 to a mature Th1-governed immune profile.170 Increased concentrations of Th2 cytokines and IgE in AYAHL subjects reduced cytotoxic T-cells and NK cells in Hodgkin lymphoma172 suggest the failure to make this Th2-to-Th1 change. These data suggest the possibility that the gut microorganisms may impact AYAHL.173,174 Cozen et al. explored whether fecal microbial diversity varied between AYAHL survivors and co-twin controls. In this small investigation, AYAHL survivors seem to have a reduction of rare gut microorganisms. Further study is required to clarify if decreased microbial diversity is a result of Hodgkin lymphoma, its therapy or a specific hygienic environment.173-176
About 12% of all human tumors are related to oncogenic viruses such as Epstein Barr Virus, Herpes Human Virus 8 and Human T-cell leukemia virus type 1.177 The occurrence of virus-related cancers, principally lymphomas, varies geographically and is induced by higher temperatures and environmental factors.178-201
Oxidative stress provoked by gut microorganisms can affect carcinogenesis and influence numerous pathways correlated with lymphomagenesis.195-201
Mucosal-associated lymphoid tissue (MALT) lymphomas are supposed to derive in the marginal zone and are connected with the presence of Helicobacter.202-205 This correlation was first revealed in an animal model infected with H. felis, an intimate relative to H. pylori and 154 days post-infection, 25% of mice had lymphoepithelial alterations while none of the controls did.206 An H. pylori infection was initially recognized in gerbils and displayed an augmentation in intestinal metaplasia.207 Since then, H. pylori infections have been recognized in animal models.208,209
H. pylori was classified as carcinogenic to humans (Group I carcinogen) in 1994 by an International Agency for Research on Cancer (IARC) Working Group based on the data from a small number of papers that studied gastric carcinoma.210 In 2009, a new Working Group evaluated significantly more results and confirmed that chronic infection with H. pylori is a Group 1 carcinogen with appropriate evidence that the infection causes gastric carcinoma and low-grade B-cell gastric MALT lymphoma.211
Gastric MALT lymphoma (GML) is strictly associated with H. pylori infection. In a retrospective evaluation of 144 consecutive patients admitted with GML, eradication treatment was extremely effective in causing complete remission (CR) and long-term prognosis was satisfactory. At multidisciplinary care stage EI, 92% of subjects received an H. pylori eradication therapy; 83% achieved CR after a mean period of seven months, and 86% remained in CR after a mean follow-up time of 105 months.212
A correlation was also hypothesized for other types of lymphomas. Numerous works described that most early-stage gastric diffuse large B-cell lymphoma (DLCBL) is H. pylori-dependent. Notably, DLCBL could possibly be treated by H. pylori eradication. Unlike MALT lymphoma, however, DLCBL may rapidly increase if it is unresponsive to H. pylori eradication. Consequently, detecting biomarkers that may predict an H. pylori-dependent status of gastric DLCBL is indispensable. Kuo et al. from Taiwan proposed that the expression of cytotoxin-associated gene A (Cag A) and CagA-signaling molecules p-SHP2 and p-ERK in malignant B cells is associated with H. pylori dependence.213 The same authors demonstrated that activating the B-cell-activating factor (BAFF) pathway upregulates NF-kB and causes BCL3 and BCL10 nuclear translocation in H. pylori-independent gastric DLCBL tumors with evidence of MALT. Moreover, they showed that the autocrine BAFF signal transduction pathway contributed to H. pylori independence in gastric MALT lymphomas without the t.11;18q21;q21 translocation.214
H. helmanii also contribute to MALT lymphoma which is preceded by endothelial venule-like vesicles, which are connected with lymphocyte enrollments.215 These models of microorganism-induced lymphoma however, appear to have variable results and may implicate bacterial and host elements.216,217
Other microorganisms such as Chlamydia psitacci, Campylobacter jejuni and Borrelia bergdorferi may also have an increase in the incidence of the disease in lymphoma progress.218
Infection of Borrelia burgdorferi may be causally related to B-cell non-Hodgkin lymphoma, as reported in one of two reports in Scandinavia.219,220Chlamydia psittaci, the agent of the zoonotic infectious disease psittacosis, has been found in MALT lymphomas in several non-gastrointestinal structures,221 while Streptococcus bovis has been connected with hematopoietic diseases such as chronic myelogenous leukemia, and chronic lymphocytic leukemia.222
Intriguing results originated from animals defective in the Ataxia telangiectasia-mutated gene (Atm-/- mice), which exhibit a high occurrence of thymic lymphoma.223 They are hypersensitive to modifications in microorganism content.224 Barlow et al. discovered that Atm-/- animals that were moved into more sterile situations lived longer and have a reduced lymphoma penetrance. In contrast, when they were relocated to standard specific-pathogenfree situations, their life and tumor latency decreased.225
Chronic Lymphocyte Leukemia (CLL)
A recent study in CLL subjects connected the efficiency of antineoplastic therapy with the use of antibiotics that alter intestinal microbiota. Pflug et al. assessed the effect of antibiotics on progression-free survival (PFS) and overall survival (OS).226 Among 800 CLL subjects, those receiving anti-Gram-positive antibiotics attained a substantially lower overall response rate (ORR). In the same study, authors evaluated patients with relapsed lymphoma. Of 122 patients with relapsed lymphoma, those treated with anti-Gram-positive antibiotics achieved a significantly lower ORR. Patients with anti-Grampositive antibiotics progressed significantly earlier than others. The multivariate analysis demonstrated that the use of anti-Gram-positive antibiotics was independently associated with reduced PFS and OS.226
More than 30% of CLLs can be classified based on their expression of stereotypic B-cell receptors (BCRs), strongly proposing that specific antigens are implicated in the onset of CLL. Unmutated CLLs, containing Ig heavy chain variable (IGHV) genes in germline configuration express low-affinity, poly- and self-reactive BCRs. Nevertheless, the antigenic specificity of CLLs with mutated IGHV-genes (M-CLL) is still elusive. In a study, Hogeeboom et al. reported a new subset of M-CLL, presenting stereotypic BCRs highly specific for β-(1,6)-glucan, a major antigenic determinant of yeasts and filamentous fungi. β-(1,6)-glucan binding depended on both the stereotypic Ig heavy and light chains, as well as on a definite amino acid in the IGHV-CDR3. Reversion of IGHV mutations to germline configuration decreased the affinity for β-(1,6)-glucan, suggesting that these BCRs are really affinity-selected for their cognate antigen. Moreover, CLL cells presenting these stereotypic receptors grow in response to β-(1,6)-glucan. With this data it is attracting to hypothesize on the possibilities for pathogen-targeted treatments for this group of subjects.227
CONCLUSION
The microbiome is currently accepted as a specific organ with separate metabolic abilities that surpass the liver’s metabolism by a factor of 100. The microbiome is able to influence hematologic malignancies via several ways, including directly through metabolites and toxins, or indirectly via the innate and adaptive immune system.228 However, a number of issues remain unresolved and only further research will clarify whether it is sufficient to administer a single species of bacteria to achieve results or whether it is better to give a mixture of microorganisms, or if by modifying an individual's microbial composition, we can improve the effectiveness of immunotherapy.229-230 In addition, FMT could help manage critical illness such as acute leukemias. In fact, in the critical care setting, several elements such as use of antibiotics, aberrant nutrition, bloodstream infections, bowel ischemia and abnormal bowel motility, strongly contribute to intestinal dysbiosis, and FMT therapy should be investigated.231,232 Further studies are needed to clarify the rationale of FMT for cancer management such as reconstruction of intestinal microbiota, amelioration of bile acid metabolism and modulation of immunotherapy efficacy.
Substances with probiotic and prebiotic capacities may represent a novel approach to change microbiota structure with beneficial effects on tumor development. Targeted treatment on the microbiome by pre-or probiotics may be used for tumor prevention and particular alterations of the microbiome may be implemented as an adjuvant treatment to augment the effectiveness of current tumor therapies of chemotherapy and immuno-therapy.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Clarissa Novello of the University of Aberdeen for the editing of the text.
DISCLOSURES
All authors declare no conflicts of interest. No funding or financial support was received.
REFERENCES
- Cho I, Blaser MJ. The human microbiome: At the interface of health and disease. Nat Rev Genet. 2012; 13:260-270.
- Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324-35.
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Ann Rev Microbiol. 2011;65:411-29.
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327-36.
- Mcdermott AJ, Huffnagle GB. The microbiome and regulation of mucosal immunity. Immunology. 2014;142:24-31.
- Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031-64.
- Sonowal R, Swimm A, Sahoo A, et al. Indoles from commensal bacteria extend healthspan. Proc Natl Acad Sci USA. 2017;114:E7506-15.
- Buddington RK, Sangild PT. Companion animals symposium: Development of the mammalian gastrointestinal tract, the resident microbiota, and the role of diet in early life. J Anim Sci. 2011;89:1506-19.
- Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735-44.
- Hansen J, Gulati A, Sartor RB. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr Opin Gastroenterol. 2010;26:564-71.
- Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunology. 2013;14:685-90.
- Musso G, Gambino R, Cassader M. Gut microbiota as a regulator of energy homeostasis and ectopic fat deposition: mechanisms and implications for metabolic disorders. Curr Opin Lipidol. 2010;21:76-83.
- Tanoue T, Honda K. Induction of Treg cells in the mouse colonic mucosa: A central mechanism to maintain host-microbiota homeostasis. Semin Immunol. 2012;24:50-7.
- Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci. 2010;107:18933-8.
- Goodrich JK, Waters JL, Poole AC, et al. Human Genetics Shape the Gut Microbiome. Cell. 2014;159:789-99.
- Cox LM, Yamanishi S, Sohn J, et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell. 2014;158:705-21.
- Jeffery IB, Lynch DB, O’Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016;10:170-82.
- Prince AL, Antony KM, Ma J, et al. The microbiome and development: a mother's perspective. Semin Reprod Med. 2014;32:1422.
- Nelson-Filho P, Borba IG, Mesquita KS, Silva RA, Queiroz AM, Silva LA. Dynamics of microbial colonization of the oral cavity in newborns. Braz Dent J. 2013;24:4159.
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:20714.
- Singh RK, Chang HW, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73.
- Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105-8.
- Reddy BS, Weisburger JH, Wynder EL. Effects of high risk and low risk diets for colon carcinogenesis on fecal microflora and steroids in man. J Nutr. 1975;105:878-84.
- Thaiss CA, Zeevi D, Levy M, et al. Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell. 2014;159:514-29.
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-63.
- Brotman RM, Shardell MD, Gajer P, et al. Association between the vaginal microbiota, menopause status, and signs of vulvovaginal atrophy. Menopause. 2014;21:450-8.
- Voigt RM, Forsyth CB, Green SJ, et al. Circadian disorganization alters intestinal microbiota. PLoS ONE. 2014;9:e97500.
- Qin J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65.
- Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biology. 2015;16:276.
- Wade WG. 2013. The oral microbiome in health and disease. Pharmacol. Res. 2013;69:13743.
- Su H, Yan X, Dong Z, Chen W, Lin ZT, Hu QG. Differential roles of Porphyromonas gingivalis lipopolysaccharide and Escherichia coli lipopolysaccharide in maturation and antigen-presenting functions of dentritic cells. Eur Rev Med Pharmacol Sci. 2015;19:248292.
- Kipanyula MJ, Seke Etet PF, Vecchio L, Farahna M, Nukenine EN, Nwabo Kamdje AHl. Signaling pathways bridging microbial-triggered inflammation and cancer. Cell Signal. 2013;25:40316.
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915-20.
- Riley DR, Sieber KB, Robinson KM, et al. Bacteria-human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Comput Biol. 2013;9:e1003107.
- Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat Chem. 2015;7:411-7.
- Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. Cytolethal distending toxin:a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology. 2011;157:1851-75.
- Lu R, Wu S, Zhang YG, et al. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis. 2014;3:e105.
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/ß-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206.
- 39. Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222-6.
- Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228-31.
- Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Ther Adv Gastroenterol. 2013;6:295-308.
- Lundin A, Bok CM, Aronsson L, et al. Gut flora, toll-like receptors and nuclear receptors: a tripartite communication that tunes innate immunity in large intestine. Cell Microbiol. 2008;10:1093-103.
- Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768-73.
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121-41.
- Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562-8.
- Vannucci L, Stepankova R, Kozakova H, Fiserova A, Rossmann P, Tlaskalova-Hogenova H. Colorectal carcinogenesis in germ-free and conventionally reared rats: different intestinal environments affect the systemic immunity. Int J Oncol. 2008;32:609-17.
- DiDonato JA, Mercurio F, Karin M. NF-?B and the link between inflammation and cancer. Immunol Rev. 2012;246:379-400.
- Musolino C, Allegra A, Innao V, Allegra AG, Pioggia G, Gangemi, S. Inflammatory and Anti-Inflammatory Equilibrium, Proliferative and Antiproliferative Balance: The Role of Cytokines in Multiple Myeloma. Mediators Inflamm. 2017;2017:1852517.
- Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736-46.
- Li D. Diabetes and pancreatic cancer. Mol Carcinog. 2012;51:64-74.
- Kuroiwa-Trzmielina J, Hesson LB. Epigenetic effects of gut microbiota on obesity and gastrointestinal cancers. In: Berger NA, editor. Epigenetics, Energy Balance, and Cancer. (Chap. 11), Switzerland: Springer International Publishing. 2016;167-89.
- Krautkramer KA, Kreznar JH, Romano KA, et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2016;64:982-92.
- Luo A, Leach ST, Barres R, et al. The Microbiota and epigenetic Regulation of T Helper 17/Regulatory T Cells:in Search of a Balanced immune System. Front Immunol. 2017;8:417.
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677-89.
- Sonnenburg, JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:doi:10.1371/journal.pbio.0040413.
- Remely M, Aumueller E, Merold C, et al. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene. 2014;537:85-92.
- Remely M, Aumueller E, Jahn D, Hippe B, Brath H, Haslberger AG. Microbiota and epigenetic regulation of inflammatory mediators in type 2 diabetes and obesity. Benef Microbes. 2014;5:33-43.
- Allegra A, Innao V, Gerace D, Bianco O, Musolino C. The metabolomic signature of hematologic malignancies. Leuk Res. 2016;49:22-35.
- Rappaport SM. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol. 2011;21:5-9.
- Imbesi S, Musolino C, Allegra A, et al. Oxidative stress in Onco-hematologic diseases: an update. Exp Rev in Hematol. 2013;6:317-25.
- Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317-28.
- Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661-72.
- Roberts AB, Wallace BD, Venkatesh MK, Mani S, Redinbo MR. Molecular insights into microbial ?-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol Pharmacol. 2013;84:208-17.
- Gill CI, Rowland IR. Diet and cancer: assessing the risk. Br J Nutr. 2002;88:S73-87.
- Loh YH, Jakszyn P, Luben RN, Mulligan AA, Mitrou PN, Khaw KT. N-Nitroso compounds and cancer incidence: the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am J Clin Nutr. 2011;93:1053-61.
- Russell WR, Hoyles L, Flint HJ, Dumas ME. Colonic bacterial metabolites and human health. Curr Opin Microbiol 2013;16:246-254.
- Pegg AE. Toxicity of polyamines and their metabolic products. Chem Res Toxicol. 2013;26:1782-800.
- Croswell A, Amir E, Teggatz P, et al. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741-53.
- Holler E, Butzhammer P, Schmid K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20:640-5.
- Galloway-Peña JR, Smith DP, Sahasrabhojane P, et al. Characterization of oral and gut microbiome temporal variability in hospitalized cancer patients. Genome Medicine. 2017;9:21.
- Lehouritis P, Cummins J, Stanton M, et al. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci Rep. 2015;5:14554.
- Selwyn FP, Cui JY, Klaassen CD. RNA-Seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab Dispos. 2015;43:1572-80.
- Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971-6.
- Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967-70.
- Gui QF, Lu HF, Zhang CX, Xu ZR, Yang, YH. Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet Mol Res. 2015;14:5642-51.
- Chitapanarux I, Chitapanarux T, Traisathit P, Kudumpee S, Tharavichitkul E, Lorvidhaya V. Randomized controlled trial of live Lactobacillus acidophilus plus Bifidobacterium bifidum in prophylaxis of diarrhea during radiotherapy in cervical cancer patients. Radiat Oncol. 2010;5:31.
- Wang Y, Luo X, Pan H, et al. Pharmacological inhibition of NADPH oxidase protects against cisplatin induced nephrotoxicity in mice by two step mechanism. Food Chem Toxicol. 2015;83:251-60.
- Mercado-Lubo R, McCormick BA. The interaction of gut microbes with host ABC transporters. Gut Microbes. 2010;1:301-6.
- West NR, Powrie F. Immunotherapy not working? Check your microbiota. Cancer Cell. 2015;28:687-9.
- Vetizou M, Pitt JM, Daillere R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-84.
- Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-1089.
- Pitt JM, Vetizou M, Boneca IG, et al. Enhancing the clinical coverage and anticancer efficacy of immune checkpoint blockade through manipulation of the gut microbiota. Oncoimmunology 2017;6:1,e1132137.
- Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-7.
- Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97-103.
- Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014;124:1174-82.
- Jenq RR, Ubeda C, Taur Y, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209:903-11.
- Jenq R, Taur Y, Devlin S, et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood and marrow Transplant 2015;21:1373-83;11.
- Hiergeist A, Koestler J, Gessner A, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood 2015;126:1723-8.
- Peled JU, Hanash AM, Jenq RR. Role of the intestinal mucosa in acute gastrointestinal GVHD. Blood. 2016;128:2395-402.
- Staffas A, Burgos da Silva M, van den Brink MR. The intestinal microbiota in allogeneic hematopoietic cell transplant and graft-versus-host disease. Blood. 2017;129:927-33.
- Mathewson ND, Reddy P. The microbiome and graft versus host disease. Curr Stem Cell Rep. 2015;1:39-47.
- Gerbitz A, Schultz M, Wilke A, et al. Probiotic effects on experimental graft-versus-host disease: let them eat yogurt. Blood. 2004;103:4365-7.
- Ladas EJ, Bhatia M, Chen L, et. al. The safety and feasibility of probiotics in children and adolescents undergoing hematopoietic cell transplantation. Bone Marrow Transplant. 2016;51:262-6.
- Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174-825.
- Jenq RR, Taur Y, Devlin SM, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21:1373-83.
- Peled JU, Devlin SM, Staffas A, et al. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol. 2017;35:1650-9.
- DeFilipp Z, Peled JU, Li S, Mahabamunuge J, et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv. 2018;2:745-53.
- Kakihana K, Fujioka Y, Suda W, et al. Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood. 2016;128:2083-8.
- Montassier E, Al-Ghalith GA, Ward T, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Medicine. 2016;8:49.
- Keefe DM, Schubert MM, Elting LS, et al. Updated clinical practice guidelines for the prevention and treatment of mucositis. Cancer. 2007;109:820-31.
- Sonis ST. The pathobiology of mucositis. Nat Rev Cancer. 2004;4:277-84.
- Berg RD. Bacterial translocation from the gastrointestinal tract. Adv Exp Med Biol. 1999;473:11-30.
- Boursi B, Mamtani R, Haynes K, et al. Recurrent antibiotic exposure may promote cancer formation - another step in understanding the role of the human microbiota? Eur J Cancer. 2015;51:2655-64.
- Friedman GD, Oestreicher N, Chan J, et al. Antibiotics and risk of breast cancer: up to 9 years of follow-up of 2.1 million women. Cancer Epidemiol Biomarkers Prev. 2006;15:2102-6.
- Rossini A, Rumio C, Sfondrini L, et al. Influence of antibiotic treatment on breast carcinoma development in proto-neu transgenic mice. Cancer Res. 2006;66:6219-24.
- Cheng M, Qian L, Shen G, et al. Microbiota modulate tumoral immune surveillance in lung through a gdT17 immune cell-dependent mechanism. Cancer Res. 2014;74:4030-41.
- Von Schwartzenberg JR, Siri TPJ. What should I eat? Cell. 2015;163:1051-2.
- Siddharth J, Holway N, Parkinson SJ. A western diet ecological module identified from the “humanized” mouse microbiota predicts diet in adults and formula feeding in children. PLoS ONE. 2013;8:e83689.
- American Cancer Society. Global Cancer Facts and Figures [Internet]. 2011 [Date accessed July 11, 2017]. Available from: http://www.cancer.org/ acs/groups/content/ epidemiologysurveilance/documents/document/ acspc-027766.pdf.
- Wu X, Patterson S, Hawk E. Chemoprevention-history and general principles. Best Pract Res Clin Gastroenterol. 2011a;25:445-59.
- Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12:5-9.
- Anhê FF, Desjardins Y, Pilona G, et al. Polyphenols and type 2 diabetes: A prospective review. Pharma Nutrition 2013;1:105-14.
- Chan AT, Giovannucci EL. Primary prevention of colorectal cancer. Gastroenterology. 2010;138:2029-43.
- Greenwald P. Cancer prevention clinical trials. J Clin Oncol. 2002;20:14-22.
- Kucuk O. New opportunities in chemoprevention. Cancer Invest. 2002;20:237-45.
- Zaibi MS, Stocker CJ, O'Dowd J, et al. Roles of GPR41 and GPR43 in leptin secretory responses of murine adipocytes to short chain fatty acids. FEBS Lett. 2010;584:2381-6.
- Peng L, Li Z-R, Green RS, et al. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619-25.
- Vinolo MA, Rodrigues HG, Nachbar RT, et al. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3:858-76.
- Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrateproducing bacteria revealed by analysis of the butyryl-CoA: acetate CoA-transferase gene. Environ Microbiol. 2010;12:304-14.
- Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: chemistry, bioavailability and effects on health. Nat Prod Rep. 2009;26:1001-43.
- Clifford MN. Diet-derived phenols in plasma and tissues and their implications for health. Planta Med. 2004;70:1103-14.
- Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem. 2013;24:1415-22.
- Etxeberria U, Fernández-Quintela A, Milagro FI, Aguirre L, Martínez JA, Portillo MP. Impact of polyphenols and polyphenol-rich dietary sources on gut microbiota composition. J Agric Food Chem. 2013;61:9517-33.
- Brown CT, Davis-Richardson AG, Giongo A, Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE 2011;6:e25792.
- Reid G, Jass J, Sebulsky MT, McCormick JK. Potential uses of probiotics in clinical practice. Clin Microb Rev. 2003;16:658-72.
- Gill HS, Cross ML, Rutherfurd KJ, Gopal PK. Dietary probiotic supplementation to enhance cellular immunity in the elderly. Br J Biomed Sci. 2001;57:94-6.
- Ogawa T, Hashikawa S, Asai Y, Sakamoto H, Yasuda K, Makimura Y. A new symbiotic, Lactobacillus casei subsp. casei together with dextran, reduces murine and human allergic reaction. FEMS Immunol Med Microbiol. 2006;46:400-9.
- Jungersen M, Wind A, Johansen E, Christensen JE, Stuer-Lauridsen B, Eskesen D. The Science behind the Probiotic Strain Bifidobacterium animalis subsp. lactis BB-12®. Microorganisms. 2014;2:92-110.
- Delcenserie V, Martel D, Lamoureux M, Amiot J, Boutin Y, Roy D. Immuno-modulatory Effects of Probiotics in the Intestinal Tract V. Curr Issues Mol Biol. 2008;10:37-54.
- Ashraf A, Mahboob S, Al-Ghanim K, et al. Immunogenic activity of lipopolysaccharides from pasteurella multocida in rabbits. J Anim Plant Sci. 2014;24:1780-5.
- Nagpal R, Kumar A, Kumar M, et al. Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiology Letters. 2012;334:1-15.
- Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7:503-14.
- Kwon HK, Lee CG, So JS, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA. 2010;107:2159-64.
- Andrade MER, Araújo RS, de Barros PAV, et al. The role of immunomodulators on intestinal barrier homeostasis in experimental models. Clin Nutr. 2015;34:1080-7.
- Tilg H, Moschen AR. Food, immunity, and the microbiome. Gastroenterology. 2015;148:1107-1119.
- Furet JP, Kong LC, Tap J, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049-57.
- Morita H, He F, Fuse T, et al. Adhesion of lactic acid bacteria to Caco-2 cells and their effect on cytokine secretion. Microbiol Immunol. 2002;46:293-7.
- Heyman M, Ménard S. Probiotic microorganisms:How they affect intestinal pathophysiology. Cell Mol Life Sci. 2002;59:1151-65.
- Zhang Y, Ma C, Zhao J, Xu H, Hou Q, Zhang H. Lactobacillus casei and vitamin K2 prevent intestinal tumorigenesis in mice via adiponectinelevated different signaling pathways. Oncotarget. 2017;8:24719-27.
- Li J, Sung CYJ, Lee N, et al. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Nat Acad Sci USA 2016;113:E1306-15.
- Rajagopala SV, Yooseph S, Harkins et al. Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis. BMC Genomics. 2016;17:635.
- Garrett WS, Lord GM, Punit S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33-45.
- Barrett AP. Leukemic cell infiltration of the gingivae. J Periodontol. 1986;57:579-81.
- Hou GL, Huang JS, Tsai CC. Analysis of oral manifestations of leukemia: a retrospective study. Oral Dis. 1997;3:31-8.
- Meyer U, Kleinheinz J, Handschel J, Kruse-Lösler B, Weingart D, Joos U. Oral findings in three different groups of immunocompromised patients. J Oral Pathol Med. 2000;29:153-8.
- Javed F, Utreja A, Bello Correa FO, et al. Oral health status in children with acute lymphoblastic leukemia. Crit Rev Oncol Hematol. 2012;83:303-9.
- Paunica SC, Dumitriu A, Mogos M, Georgescu O, Mogos I. The evaluation of the periodontium in patients with leukemia using thermographic imaging. Hematology. 2009;14:341-6.
- Khan SA, Wingard JR. Infection and mucosal injury in cancer treatment. JNCI Monographs. 2001;29:31-6.
- Greenberg MS, Cohen SG, McKitrick JC, Cassileth PA. The oral flora as a source of septicemia in patients with acute leukemia. Oral Surg Oral Med Oral Pathol. 1982;53:32-6.
- Sixou JL, De Medeiros-Batista O, Gandemer V, Bonnaure-Mallet M. The effect of chemotherapy on the supragingival plaque of pediatric cancer patients. Oral Oncol. 1998;34:476-83.
- Wahlin YB, Granstrom S, Persson S, Sjöström M. Multivariate study of enterobacteria and Pseudomonas in saliva of patients with acute leukemia. Oral Surg Oral Med Oral Pathol. 1991;72:300-8.
- O’Sullivan EA, Duggal MS, Bailey CC, Curzon ME, Hart P. Changes in the oral microflora during cytotoxic chemotherapy in children being treated for acute leukemia. Oral Surg Oral Med Oral Pathol. 1993;76:161-8.
- Galili D, Donitza A, Garfunkel A, Sela MN. Gram-negative enteric bacteria in the oral cavity of leukemia patients. Oral Surg Oral Med Oral Pathol. 1992;459-62.
- Wang.Y, Xue J, Zhou X, et al. Oral Microbiota Distinguishes Acute Lymphoblastic Leukemia Pediatric Hosts from Healthy Populations. PLoS One 2014;9:e102116.
- Lucas VS, Beighton D, Roberts GJ, Challacombe SJ. Changes in the oral streptococcal flora of children undergoing allogeneic bone marrow transplantation. J Infect. 1997;35:135-41.
- Phulpin-Weibel A, Gaspar N, Emirian A, Chachaty E, Valteau-Couanet D, Gachot B. Intravascular catheter-related bloodstream infection caused by Abiotrophia defectiva in a neutropenic child. J Med Microbiol. 2013;62:789-91.
- Senn L, Entenza JM, Greub G, Jaton K, Wenger A, Bille J, et al. Bloodstream and endovascular infections due to Abiotrophia defectiva and Granulicatella species. BMC Infect Dis. 2006;6:9.
- Cargill JS, Scott KS, Gascoyne-Binzi D, Sandoe JA. Granulicatella infection: diagnosis and management. J Med Microbiol. 2012;61:755-61.
- Liao CH, Teng LJ, Hsueh PR, et al. Nutritionally variant streptococcal infections at a University Hospital in Taiwan: disease emergence and high prevalence of beta-lactam and macrolide resistance. Clin Infect Dis. 2004;38:452-5.
- Soares AF, Aquino AR, Carvalho CH, Nonaka CF, Almeida D, Pinto LP. Frequency of oral mucositis and microbiological analysis in children with acute lymphoblastic leukemia treated with 0.12% chlorhexidine gluconate. Braz Dent J. 2011;22:3126.
- Chua LL, Rajasuriar R, Azanan MS, et al. Reduced microbial diversity in adult survivors of childhood acute lymphoblastic leukemia and microbial associations with increased immune activation. Microbiome. 2017;5:35.
- Sung L, Lange B, Gerbing R, et al. Microbiologically documented infections and infection-related mortality in children with acute myeloid leukemia. Blood. 2007;110:3532-9.
- Kachlany SC, Schwartz AB, Balashova NV, et al. Anti-leukemia activity of a bacterial toxin with natural specificity for LFA-1 on white blood cells. Leuk Res. 2010;34:777-85.
- Jalali F, Sharifi M, Salehi R. Kefir induces apoptosis and inhibits cell proliferation in human acute erythroleukemia. Med Oncol. 2016;33:7.
- Bu S, Xie Q, Chang W, Huo X, Chen F, Ma X. LukS-PV induces mitochondrial-mediated apoptosis and G 0/G 1 cell cycle arrest in human acute myeloid leukemia THP-1 cells. Int J Biochem Cell Biol. 2013;45:1531-7.
- Shan W, Bu S, Zhang C, et al. LukS-PV, a component of Panton-valentine leukocidin, exerts potent activity against acute myeloid leukemia in vitro and in vivo. Int J Biochem Cell Biol. 2015;61,20-8.
- Dai C, Zhang C, Sun X, et al. LukS-PV induces differentiation by activating the ERK signaling pathway and c-JUN/c-FOS in human acute myeloid leukemia cells. Int J Biochem Cell Biol. 2016;76:107-14.
- Shan W, Ma X, Deng F. Is LukS-PV a novel experimental therapy for leukemia? Gene. 2017;600:44-7.
- Salas C, Niembro A, Lozano V, et al. Persistent genomic instability in peripheral blood lymphocytes from Hodgkin lymphoma survivors. Environ Mol Mutagen. 2012;53:271-80.
- Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 2010;107:11971-75.
- Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 2011;108:4578-85.
- Martin R, Nauta AJ, Ben Amor K, Knippels LM, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benef Microbes. 2010;1:367-82.
- Poppema S. Immunobiology and pathophysiology of Hodgkin lymphomas. Hematology Am Soc Hematol Educ Prog. 2005;231-8.
- Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv7.
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268-73.
- Cozen W, Yu G, Gail MH. Fecal microbiota diversity in survivors of adolescent/young adult Hodgkin lymphoma:a study of twins. Br J Cancer 2013;108:1163-7.
- Parkin, Donald Maxwell. The global health burden of infection-associated cancers in the year Int. J Cancer. 2002;118:3030.
- IARC Working Group on the Evaluation of Carcinogenic Risk to Humans. Epstein-Barr Virus and Kaposi's Sarcoma Herpesvirus/Human Herpesvirus 8. Lyon (FR): International Agency for Research on Cancer; 1997 [Date accessed May 20, 2017]. (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, No. 70.) Available from: https://www. ncbi.nlm.nih.gov/books/NBK385507/
- Campo E, Swerdlow SH, Harris NL, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117:5019-5032.
- Cesarman E, Spina M, Gloghini A, et al. HIV-associated lymphomas and gamma herpesviruses. Blood. 2009;113:1213-24.
- Chadburn A, Chiu A, Lee JY, et al. Immunophenotypic analysis of AIDS-related diffuse large B-cell lymphoma and clinical implications in patients from AIDS Malignancies Consortium clinical trials 010 and 034. J Clin Oncol. 2009;27:5039-48.
- Ghobrial IM, Habermann TM, Maurer MJ, et al. Prognostic analysis for survival in adult solid organ transplant recipients with post-transplantation lymphoproliferative disorders. J Clin Oncol. 2005;23:7574-82.
- Nelson BP, Nalesnik MA, Bahler DW, et al. Epstein-Barr virus-negative post-transplant lymphoproliferative disorders: A distinct entity? Am J Surg Pathol. 2000;24:375-85.
- Araujo I, Foss HD, Bittencourt A, et al. Expression of Epstein-Barr virus-gene products in Burkitt’s lymphoma in Northeast Brazil. Blood. 1996;87:5279-86.
- Glaser SL, Lin RJ, Stewart SL, et al. Epstein-Barr virus-associated Hodgkin’s disease: epidemiologic characteristics in international data. Int J Cancer. 1997;70:375-82.
- Brown HJ, Song MJ, Deng H, et al. NF-kappaB inhibits gammaherpesvirus lytic replication. J Virol. 2003;77:8532-40.
- Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalised lymphocytes transforms established rodent cells. Cell. 1985;43:831-40.
- Huen DS, Henderson SA, Croom-Carter, et al. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549-60.
- Arvanitakis L, Yaseen N, Sharma S. Latent membrane protein-1 induces cyclin D2 expression, pRb hyperphosphorylation, and loss of TGF-beta 1-mediated growth inhibition in EBV-positive B cells. J Immunol. 1995;155:1047-56.
- Moore PS, Chang Y. Molecular virology of Kaposi’s sarcoma-associated herpesvirus. Philos Trans R Soc Lond B Biol Sci. 2001;356499-516.
- Deacon EM, Pallesen G, Niedobitek G, et al. Epstein-Barr virus and Hodgkin’s disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med. 1993;177:339-49.
- Caldwell RG, Wilson JB, Anderson SJ, et al. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405-11.
- Sharp TV, Schwemmle M, Jeffrey I, et al. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483.
- Ramos JC, Lossos IS. Newly emerging therapies targeting viral-related lymphomas. Curr Oncol Rep. 2011;13:416-26.
- Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2:920-32.
- Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341-6.
- Oliver AM, Martin F, Kearney JF. IgMhighCD21high lymphocytes enriched in the splenic marginal zone generate effector cells more rapidly than the bulk of follicular B cells. J Immunol. 1999;162:7198-207.
- Kullisaar T, Zilmer M, Mikelsaar M. Two antioxidative lactobacilli strains as promising probiotics. Int J Food Microbiol. 2002;72:215-24.
- Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. 2007;121:2381-6.
- Epeldegui M, Widney DP, Martinez-Maza O. Pathogenesis of AIDS lymphoma:role of oncogenic viruses and B cell activation-associated molecular lesions. Curr Opin Oncol. 2006;18:444-8.
- Illes A, Varoczy L, Papp G, et al. Aspects of B-cell non-Hodgkin’s lymphoma development: a transition from immune-reactivity to malignancy. Scand J Immunol. 2009;69:387-400.
- Saito Y, Suzuki H, Tsugawa H, et al. Overexpression of miR-142-5p and miR-155 in gastric mucosa-associated lymphoid tissue.MALT] lymphoma resistant to Helicobacter pylori eradication. PLoS One 2012;7:doi:10.1371/ journal.pone.0047396.
- Isaacson PG, Du MQ. MALT lymphoma: from morphology to molecules. Nat Rev Cancer. 2004;4:644-53.
- Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338;1175-6.
- Bayerdorffer E, Rudolph B, Neubauer A, et al. Malt Lyphoma Study Group. Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. MALT Lymphoma Study Group. Lancet. 1995;345:1591-4.
- Enno A, O'Rourke JL, Howlett CR, Jack A, Dixon MF, Lee A. MALToma-like lesions in the murine gastric mucosa after long-term infection with Helicobacter felis. A mouse model of Helicobacter pylori-induced gastric lymphoma. Am J Pathol. 1995;147:217-22.
- Hirayama F, Takagi S, Kusuhara H, Iwao E, Yokoyama Y, Ikeda Y. Induction of gastric ulcer and intestinal metaplasia in Mongolian gerbils infected with Helicobacter pylori. J Gastroenterol. 1996;31:755-7.
- Mueller A, O'Rourke J, Grimm J. Distinct gene expression profiles characterize the histopathological stages of disease in Helicobacter-induced mucosa-associated lymphoid tissue lymphoma. Proc Natl Acad Sci. USA 2003;100:1292-7.
- O’Rourke JL. Gene expression profiling in Helicobacter-induced MALT lymphoma with reference to antigen drive and protective immunization. J Gastroenterol Hepatol. 2008;23:151-6.
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Schistosomes, Liver Flukes and Helicobacter pylori. 1994 [Date accessed March 22, 2018]. Available from: https://monographs.iarc.fr/ENG/ Monographs/vol61/mono61.pdf
- ARC Working Group on the Evaluation of Carcinogenic Risks to Humans. A Review of Human Carcinogens. Part B: Biological Agents. 2012 [Date accessed March 22, 2018]. Available from: http://monographs.iarc.fr/ ENG/Monographs/vol100B/mono100B.pdf.
- Moleiro J, Ferreira S, Lage P, Dias Pereira A. Gastric malt lymphoma: Analysis of a series of consecutive patients over 20 years. United European Gastroenterol J. 2016;4:395-402.
- Kuo SH, Chen LT, Lin CW, et al. Expressions of the CagA protein and CagA- signaling molecules predict Helicobacter pylori dependence of early-stage gastric DLBCL. Blood. 2016;129:188-98.
- Kuo SH, Tsai HJ, Lin CW, et al. The B-cell-activating factor signaling pathway is associated with Helicobacter pylori I dependence in gastric mucosa-associated lymphoid tissue lymphoma without t(11;18)(q21;q21). J Pathol. 2017;241:420-33.
- Suzuki A, Kobayashi M, Matsuda K, et al. Induction of high endothelial venule-like vessels expressing GlcNAc6ST-1-mediated L-selectin ligand carbohydrate and mucosal addressin cell adhesion molecule 1 (MAdCAM-1) in a mouse model of “Candidatus Helicobacter heilmannii”- induced gastritis and gastric mucosa-associated lymphoid tissue (MALT) lymphoma. Helicobacter. 2010;15:538-48.
- O’Rourke JL, Dixon MF, Jack A, Enno A, Lee A. Gastric B-cell mucosa-associated lymphoid tissue (MALT) lymphoma in an animal model of “Helicobacter heilmannii” infection. J Pathol. 2004;203:896-903.
- Nakamura M, Murayama SY, Serizawa H, et al. “Candidatus Helicobacter heilmannii” from a cynomolgus monkey induces gastric mucosa-associated lymphoid tissue lymphomas in C57BL/6 mice. Infect Immun. 2007;75:1214-22.
- Suarez F, Lortholary O, Hermine O, Lecuit M. Infection-associated lymphomas derived from marginal zone B cells:a model of antigen-driven lymphoproliferation. Blood. 2006;107:3034-44.
- Schollkopf C, Melbye M, Munksgaard L, et al. Borrelia infection and risk of non-Hodgkin lymphoma. Blood. 2008;111:5524-9.
- Chang CM, Landgren O, Koshiol J, et al. Borrelia and subsequent risk of solid tumors and hematologic malignancies in Sweden. Int J Cancer. 2012;131:2208-09.
- Aigelsreiter A, Gerlza T, Deutsch AJ, et al. Chlamydia psittaci infection in nongastrointestinal extranodal MALT lymphomas and their precursor lesions. Am J Clin Pathol. 2011;135:70-5.
- Gold JS, Bayar S, Salem RR. Association of Streptococcus bovis bacteremia with colonic neoplasia and extracolonic malignancy. Arch Surg. 2004;139:760-5.
- Barlow C, Hirotsune S, Paylor R, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 1996;86:159-71.
- Reliene R, Schiestl RH. Antioxidant N-acetyl cysteine reduces incidence and multiplicity of lymphoma in Atm deficient mice. DNA Repair. 2006;5:852-9.
- Pflug N, Kluth S, Vehreschild JJ, et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology. 2016;5:e1150399.
- Hoogeboom R, van Kessel KP, Hochstenbach F, et al. A mutated B cell chronic lymphocytic leukemia subset that recognizes and responds to fungi. J Exp Med. 2013;210:59-70
- Goodman B, Gardner H. The microbiome and cancer. J Pathol. 2018;244:667-76.
- Rescigno M. A ‘fit’ microbiota to potentiate cancer immunotherapy. Genome Medicine. 2015;7:131.
- Baquero F, Nombela C. The microbiome as a human organ. Clin Microbiol Infect. 2012;18:2-4.
- Limketkai BN, Hendler S, Ting PS, Parian AM. Fecal Microbiota Transplantation for the Critically Ill Patient. Nutr Clin Pract. 2019;34:73-9.
- Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management:Current status and perspectives. Int J Cancer. 2018. doi:10.1002/ijc.32003.




